At the ”omic” level, CSCs are different than their non-CSC counterparts. These differences are believed to be epigenetically driven as well as a consequence of post translational modifications. We have dissected CSCs and discovered what makes then tick, and learned how to genetically and pharmacologically target these weaknesses.
Using different approaches, including high-throughput methylation analysis, mircoRNA arrays, and RNAseq, we are closer to understating what makes a CSC a CSC.
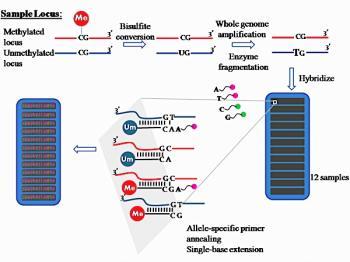

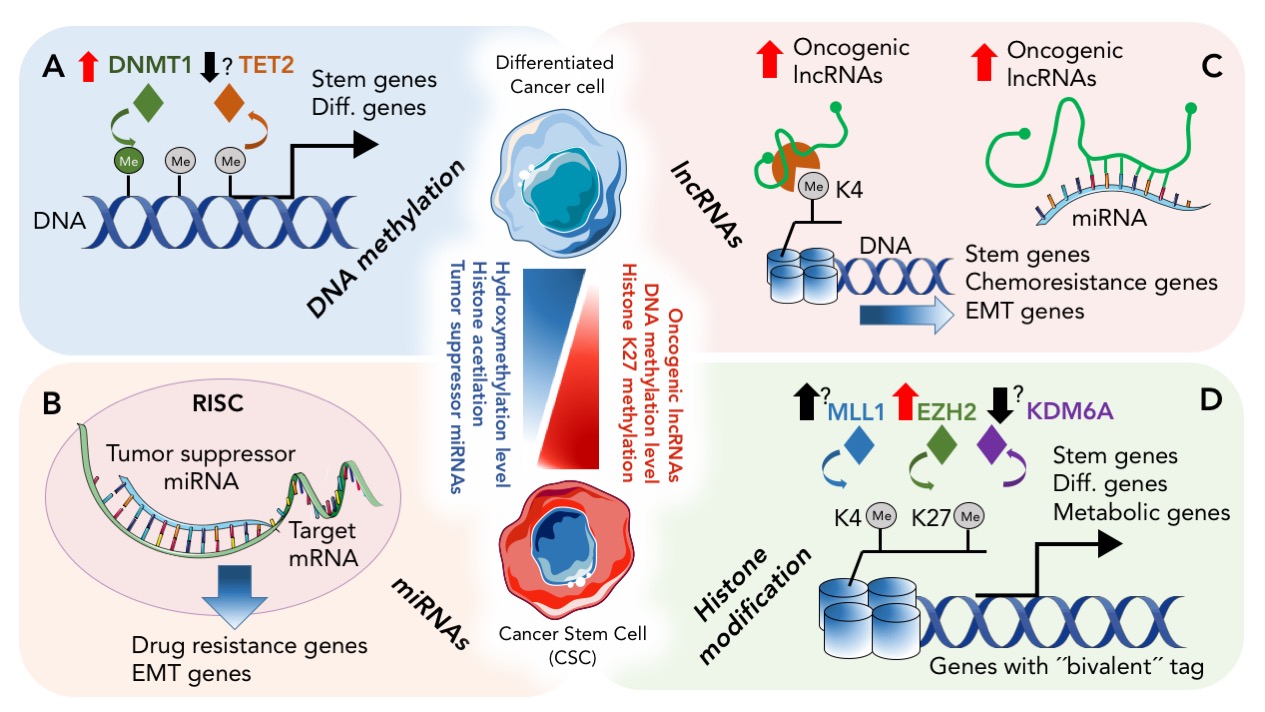 Epigenetic regulation of Cancer Stem Cells. Graphical summary of the epigenetic mechanisms regulating CSCs. DNA methylation, histone modifications and non-coding RNA molecules (lncRNAs and miRNAs) work together to modify CSC biology and plasticity. (A) The DNMT1 methyltransferase methylates CpG sites contributing to differential methylation of genes important for stemness and differentiation. It is likely that in parallel, TET proteins generate 5hmC conferring active or passive demethylation. While several studies have shown that DNMT1 is over expressed in CCS, the level of TET proteins in CSC is yet to be determined. Non-coding RNA molecules, specifically miRNA and lncRNA molecules add an additional layer of complexity and regulation. (B) miRNAs can target a number of mRNAs that play important roles as pro- or anti-CSC regulators. For example, the miR17-92 cluster targets a number of genes important for pancreatic CSC-ness, and as such the expression of this cluster is downregulated in CSCs. On the other hand, miRNAs that promote EMT features, such as the loss of cell adhesion, are upregulated in CSC. Examples include miR-9 (target: E-cadherin), miR-194 (target N-cadherin), miR-661 (target: Nestin and Star1). mRNAs important for self-renewal, EMT and chemoresistance. (C) lncRNA can represent a bridge between the many layers of epigenetic gene regulation, interacting with, for example, histone modifiers or serving as ceRNAs for miRNAs. (D) Genes important for stemness, differentiation (diff.) and metabolic reprogramming are found with active and inhibiting histone tags, and histone modifying enzymes are expressed at different levels between CSCs and their non-CSCs counterparts, such asMLL1, EZH2 and KDM6A. (Center) The plasticity of differentiated cancer cells and CSCs and their ability to quickly transdifferentiate could be a due to a balance between, high and/or low levels of DNA methylation, hydroxymethylation, histone acetylation or methylation, together with tumor-suppressive/oncogenic miRNAs and lncRNAs. Learn more
Epigenetic regulation of Cancer Stem Cells. Graphical summary of the epigenetic mechanisms regulating CSCs. DNA methylation, histone modifications and non-coding RNA molecules (lncRNAs and miRNAs) work together to modify CSC biology and plasticity. (A) The DNMT1 methyltransferase methylates CpG sites contributing to differential methylation of genes important for stemness and differentiation. It is likely that in parallel, TET proteins generate 5hmC conferring active or passive demethylation. While several studies have shown that DNMT1 is over expressed in CCS, the level of TET proteins in CSC is yet to be determined. Non-coding RNA molecules, specifically miRNA and lncRNA molecules add an additional layer of complexity and regulation. (B) miRNAs can target a number of mRNAs that play important roles as pro- or anti-CSC regulators. For example, the miR17-92 cluster targets a number of genes important for pancreatic CSC-ness, and as such the expression of this cluster is downregulated in CSCs. On the other hand, miRNAs that promote EMT features, such as the loss of cell adhesion, are upregulated in CSC. Examples include miR-9 (target: E-cadherin), miR-194 (target N-cadherin), miR-661 (target: Nestin and Star1). mRNAs important for self-renewal, EMT and chemoresistance. (C) lncRNA can represent a bridge between the many layers of epigenetic gene regulation, interacting with, for example, histone modifiers or serving as ceRNAs for miRNAs. (D) Genes important for stemness, differentiation (diff.) and metabolic reprogramming are found with active and inhibiting histone tags, and histone modifying enzymes are expressed at different levels between CSCs and their non-CSCs counterparts, such asMLL1, EZH2 and KDM6A. (Center) The plasticity of differentiated cancer cells and CSCs and their ability to quickly transdifferentiate could be a due to a balance between, high and/or low levels of DNA methylation, hydroxymethylation, histone acetylation or methylation, together with tumor-suppressive/oncogenic miRNAs and lncRNAs. Learn more
Our laboratory continues to work towards better understanding the “omic” landscape of CSCs and non-CSCs in order to design and develop new therapeutics to specifically target CSCs